We Solve medical tasks using working aptamers
Aptamer Selection, Characterization, 3D Structure Simulation And Target Identification
APTAMER DESIGN
AND 3D STRUCTURE
We will determinate of the tertiary structure and make the molecular design of your aptamers, just click the button
When tertiary structure determination and molecular aptamer design are required
IF YOU NEED TO DETERMINE THE NATIVE APTAMER CONFORMATION IN THE SOLUTION
We combine methods of computer simulation such as molecular dynamics (MD) simulations and quantum chemical calculations with the small-angle X-ray scattering (SAXS) experiment to reveal the structure of the aptamer in the solution. The developed approach allows us to choose the most probable secondary structure of the aptamer from the variety of options offered by the folding servers and obtain the aptamer conformation taking into account solution temperature, ions presence and other conditions. We use computer simulations to determine which nucleotides are responsible for the stability of the aptamer conformation.
IF YOU NEED A MOLECULAR STRUCTURE MODEL ON A COMPUTER
Computer molecular design is widely used to visualize the complex spatial structure by three-dimensional (3D) molecular models. Aptamer 3D models are applied for directed design of aptamers in order to enhance their specificity and affinity, prediction of the binding sites and explaining the mechanisms of interaction with a target.
STAGES OF MOLECULAR DESIGN
1
We determine the most probable spatial structure of the aptamer in a buffer solution using the small-angle X-ray scattering (SAXS) method.
2
We process the data of the SAXS experiment, calculate the structural parameters, estimate the molecular weight of the molecule in solution, and construct a three-dimensional electron density of the molecule from pseudoatoms.
3
Based on the aptamer sequence we perform computer molecular design aimed at obtaining the secondary structure options and respective three-dimensional models.
4
We carry out molecular dynamics (MD) simulation of the spatial structures to predict the native aptamer conformation in the solution. We compare the MD and SAXS data to find out which model fits the experimental results better.
STAGES IN MORE DETAIL
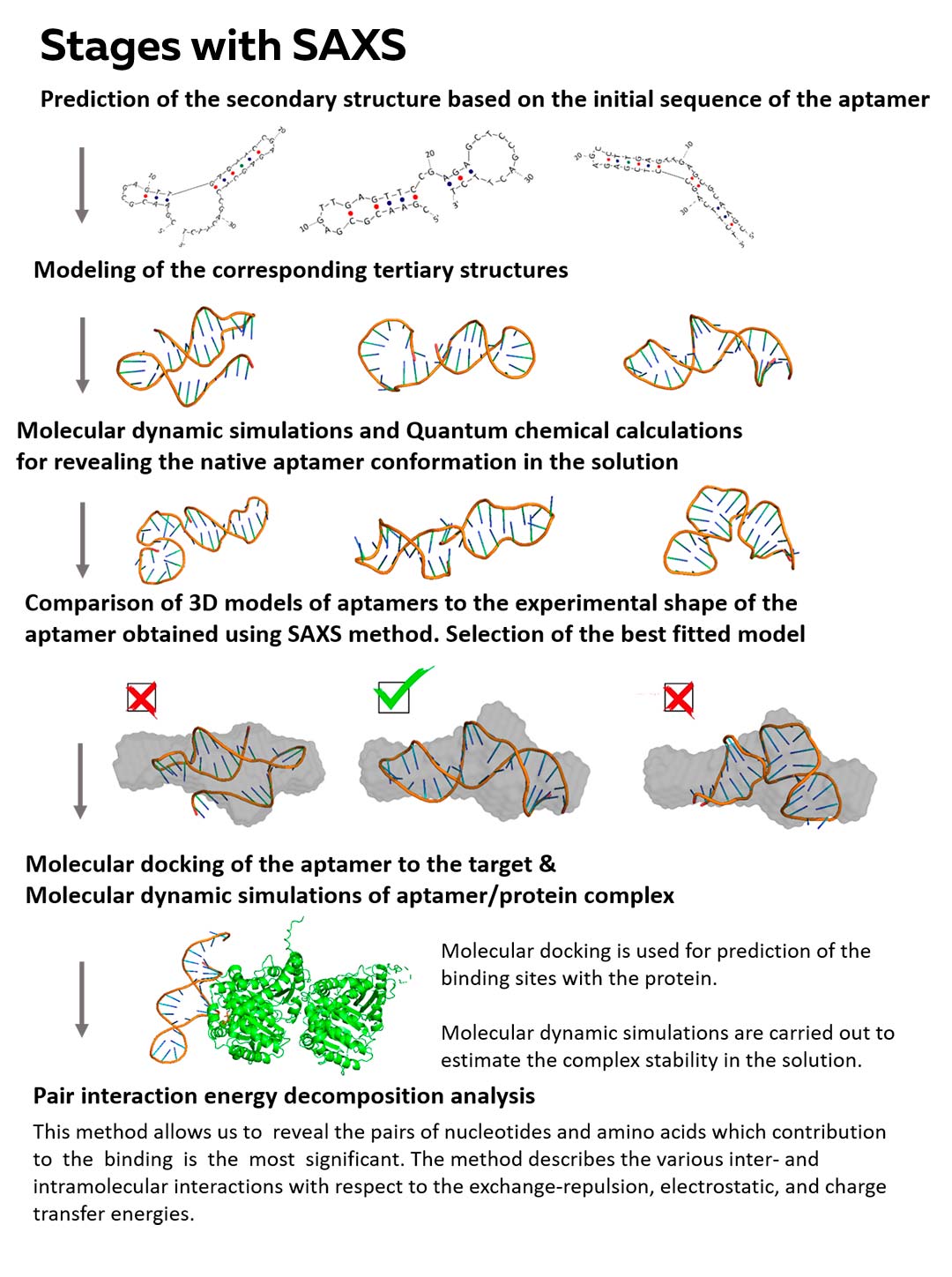
Stages with SAXS
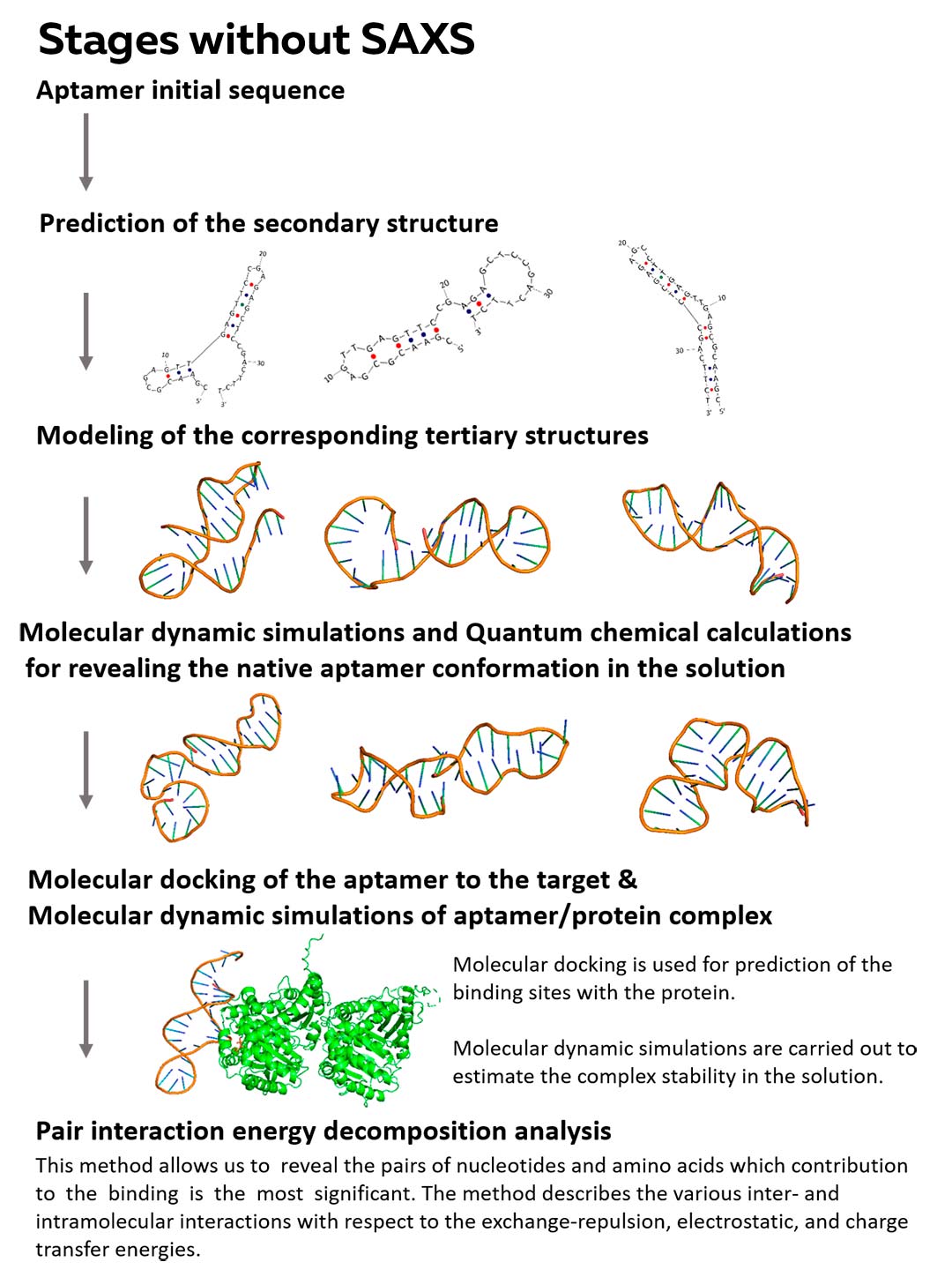
Stages without SAXS
WE USE INNOVATive TECHNOLOGIES
APTABID
AptaBiD allows us to isolate proteins that bind to specific aptamers. With the help of this method it becomes possible to determine the direct target of the aptamer.
TISSUE SELEX
The method for generating aptamers which are capable to bind with tissue targets.
CELL SELEX
This technology involves the selection of aptamers for the whole cells. This allows us to obtain aptamers that are able to bind to the target in its native conformation.
LIGS
The method identifies highly selective aptamers against a predetermined epitope expressed on the cell surface. Monoclonal antibody interacts with its cognate epitope to outcompete and replace specific aptamers from an enriched SELEX pool.
SAXS
This method is used for determination of the nanoscale density differences in the sample. This makes it possible to determine the size and shape of the macromolecule.
HT SELEX
HT SELEX is the method that increases the sensitivity over traditional cloning and sequencing applications and reduces the number of rounds required to achieve detectable enrichment.
THE RESULTS YOU'LL GET
SCIENTISTS INVOLVED IN THE STUDY

VLADIMIR ZABLUDA

POLINA ARTYUSHENKO


 .
.